QPCR Screening of Pediatric Saliva Samples to Evaluate Effects of Dental Sealants on Cariogenic Bacteria Streptococcus Mutans and Scardovia Wiggsiae
Olivia Tsang1 , Kevin Major1 , Sonia Santoyo1 , Karl Kingsley2 and Linh Nguyen2*
1Department of Clinical Sciences, University of Nevada, Las Vegas - School of Dental Medicine, Las Vegas, United States .
2Department of Biomedical Sciences, University of Nevada, Las Vegas - School of Dental Medicine, Las Vegas, United States .
Corresponding author Email: Karl.Kingsley@unlv.edu
DOI: http://dx.doi.org/10.12944/EDJ.02.01.04
Copy the following to cite this article:
Tsang O, Major K, Santoyo S, Kingsley K, Nguyen L. qPCR Screening of Pediatric Saliva Samples to Evaluate Effects of Dental Sealants on Cariogenic Bacteria Streptococcus Mutans and Scardovia Wiggsiae. Enviro Dental Journal 2021; 2(1). DOI:http://dx.doi.org/10.12944/EDJ.02.01.04
Copy the following to cite this URL:
Tsang O, Major K, Santoyo S, Kingsley K, Nguyen L. qPCR Screening of Pediatric Saliva Samples to Evaluate Effects of Dental Sealants on Cariogenic Bacteria Streptococcus Mutans and Scardovia Wiggsiae. Enviro Dental Journal 2021; 2(1). Available From : https://bit.ly/3sH7z1i
Download article (pdf) Citation Manager
Select type of program for download
| Endnote EndNote format (Mac & Win) | |
| Reference Manager Ris format (Win only) | |
| Procite Ris format (Win only) | |
| Medlars Format | |
| RefWorks Format RefWorks format (Mac & Win) | |
| BibTex Format BibTex format (Mac & Win) |
Article Publishing History
| Received: | 27-10-2019 |
|---|---|
| Accepted: | 08-01-2021 |
| Reviewed by: | Priyanka Manchanda |
| Second Review by: | Sachidanand Mallya P |
| Final Approval by: | Prof. Azmina Hussain |
Introduction
Dental caries is an alarmingly prevalent multifactorial disease worldwide but is most common as a chronic disease among children and adolescents1. Lifestyle factors such as socioeconomic status, diet, and dental knowledge are known to contribute to early childhood caries (ECC). However, biological influences such as saliva and the composition of cariogenic bacteria within the oral biome contribute largely contribute to ECC as well2,3.
Within the oral cavity there are more than 700 bacterial species, some of which are known to be associated with being cariogenic pathogens, such as Streptococcus mutans (S. mutans) and Scardoviawiggsiae (S. wiggsiae)4,5,6. Both species are acid producing and acid tolerant, which aid in the erosion of tooth enamel and provide the foundation for caries to occur7. S. mutans, in particular, has been widely studied to have a strong association with ECC but recent studies have emerged indicating the presence of S. wiggsiae in children with severe ECC8,9.
Although dental caries are highly prevalent, it is also preventable. Methods such as dental sealants or fluoride treatment, which have proven to be highly effective in reducing caries risk, are widely recommended by professional health organizations10,11. Even though there have been many studies on the presence and abundance of cariogenic pathogens, there are limited sources on the exact effect of these treatments on S. mutans and S. wiggsiaewhen preventative measures are taken12,13. Due to the lack of conclusive evidence, this study is aimed to evaluate the microbial burden of S. mutans and S. wiggsiae among pediatric patients in a dental school to analyze the effects dental sealants have on the oral microbiota in the pediatric population.
Methodology
Study Approval
The protocol for this study was eviewed by the Office for the Protection of Research Subjects (OPRS) and Institutional Review Board (IRB) OPRS#880427-1 “The Prevalence of Oral Microbes in Saliva from the University of Nevada Las Vegas (UNLV) School of Dental Medicine (SDM) pediatric and adult clinical population”.
Sample collection
The original saliva samples were collected from aa randomized pool of pediatric patients at UNLV SDM with Informed Consent from parents/guardians at the time of enrollment. Pediatric assent and Parental permission to consent for voluntary participation were both required at the time of study enrollment. Inclusion criteria included patients aged seven (7) years and older. Exclusion criteria included any child that was not a patient at UNLV SDM, any child who declined to participate, and any parent or guardian that declined participation.
In brief, pediatric patients originally enrolled in the sample collection study were given a sterile 50mL container for saliva collection, and were instructed to provide up to 5mL for analysis. Samples were stored on ice until transfer to biomedical laboratory to process and analyze results. Research bias was prevented by using a randomly, non-duplicated number which was assigned to each sample. There were no patient-specific identifying information available to research team members.
Sealant Placement Protocol
As part of the standard clinical treatment, pediatric patients received standardized dental sealants placed directly on occlusal surfaces of molars and premolars. Patients had Isolite or cotton isolation for the dental sealant placement with 3M ESPE Clinpro Sealant technique guide. Modifications were made to the placement of bonding agent between etching and placing the sealant. Each tooth receiving a dental sealant was etched for 15 seconds with 37% phosphoric acid etch, bonded with OptibondSolo Plus, sealed with 3M ESPE Clinpro Sealant, and cured for 20 seconds.
Saliva Collection
Once sealants were placed for appropriate patient treatment, the patient was scheduled for a follow up appointment according to clinic protocol. Patients were scheduled for 90 day recall appointments, with a majority returning within six months of the initial collection date. At the follow up appointment, patients followed initial collection sample protocol and provided post-sealant samples for analysis.
DNA Isolation
All previously collected samples (pre-sealant, post-sealant) were stored in a biomedical laboratory at -80°C. To screen for the presence of S. wiggsiae (SW) of S. mutans (SM), DNA was subsequently isolated from each patient sample using the GentraPuregene Blood Kit. A brief summary of this procedure follows:
|
Step 1 |
One mL of saliva concentrated by centrifugation at 5,000 revolutions per minute (RPM) for 5 minutes. |
|
Step 2 |
The supernatant was discarded and cell pellet dissolved in 275uL of cell lysis solution. |
|
Step 3 |
1.5uL Puregene Proteinase K was added and mixed by inverting 25 times. |
|
Step 4 |
Each sample was then incubated at 55°C for one hour then place at 4°C. |
|
Step 5 |
100uL of protein precipitation solution was added to each sample, vortexed for 15 seconds, incubated on ice for 10 minutes, and then centrifuged at 14,000 RPM for five minutes. |
|
Step 6 |
300 uL isopropanol was pipetted into a clean 1.5 mL tube and the supernatant from the previous step was transferred. |
|
Step 7 |
0.5 uL Glycogen Solution was then added, mixed by inverting 50 times, incubated for five minutes at room temperature, and centrifuged at 14,000 RPM for five minutes. |
|
Step 8 |
The supernatant was discarded and 300 uL of 70% ethanol was added, inverted several times to wash the DNA pellet, and then centrifuged at 14,000 RPM for five minutes. |
|
Step 9 |
The supernatant was removed and the pellet was allowed to dry for five minutes. |
|
Step 10 |
50 uL of sterile water was then added and vortexed for five seconds at medium speed. |
|
Step 11 |
Each sample was then incubated at 65°C for one hour to dissolve DNA then incubated at room temperature overnight with gentle shaking |
|
Step 12 |
The DNA isolates were subsequently stored at 4°C |
qPCR Screening
Screening for the presence of SW was accomplished using quantitative polymerase chain reaction (qPCR) with specifications that included an initial incubation at 50°C for two minutes, followed by denaturation at 95°C for 10 minutes, and 40 cycles: denaturation at 95°C for 15 seconds and annealing 51°C for one minute. Screening for the presence of SM was also accomplished using qPCR with same specifications as SW, but with the exception of the annealing temperature of 58°C. Standard curve for relative quantification of SW was constructed from a patient’s saliva sample that was previously known to contain high concentrations of SW5. The standard curve for SM was constructed from purified SM reference strains obtained from the American Type Culture Collection (ATCC-25175), which was used for absolute quantitation. The reaction included: 15 uLTaqMan universal PCR master mix (Applied Biosystems), 0.6 uL of the following primers at concentration of 10 uM from Eurofins MWG Operon (Huntsville, AL), 0.75 uL of probe resulting in final probe concentration of 0.2 uM, with 2 uL of DNA samples. Sterile, nuclease-free distilled water from Promega (Madison, WI) was used to adjust the final reaction volume to 30 uL. Each screening was performed in duplicate.
Primers synthesize from Eurofins MWG Operon (Huntsville, AL) were:
(Nucleotide = nt)
Forward 16S rRNA universal primer, 5’-ACGCGTCGACAGAGTTTGATCCTGGCT-3’; 27 nt, 56% GC, Tm 76°C
Reverse 16S rRNA universal primer, 5’-GGGACTACCAGGGTATCTAAT-3’; 21 nt, 48% GC, Tm 62°C
Annealing temperature: 63°C
Optimal temperature T(opt): Lower temperature – 5°C = 58°C
Forward primer-SW, 5’-GTGGACTTTATGAATAAGC-3’; 19 nt, 37% GC, Tm 55°C
Reverse primer- SW, 5’-CTACCGTTAAGCAGTAAG-3’; 18 nt, 44% GC, Tm 56°C
Annealing temperature: 56°C
Optimal temperature T(opt): Lower temperature – 5°C = 50°C
Forward primer-SM, 5’-GCCTACAGCTCAGAGATGCTATTCT-3’; 25 nt, 48%GC, Tm 68°C
Reverse primer-SM, 5’- GCCATACACCACTCATGAATTGA-3’; 23 nt, 43% GC, Tm 65°C
Optimal temperature T(opt): Lower temperature – 5°C = 60°C
Statistical Analysis
The analysis used for this experiment was Pearson’s Chi squared test, which is appropriate for studies using non-parametric data with smaller sample sizes. Categorical non-parametric data included qPCR screening results for both pre- and post- sealant samples,with an overall sample size (n=26).
Results
Basic demographic information was collected and summarized for the samples analyzed in this study (Table 1). In brief, equal numbers of femalesand males were represented in the study sample, which is not significantly different from the overall clinic population (p=0.8414). The majority of the patients were non-White (Hispanic) minorities, which was also similar to the overall pediatric clinic population (p=0.2225).
Table 1: Demographic Analysis of Study Sample
|
|
Study sample (n=26) |
Pediatric clinic |
Statistical analysis |
|
Sex |
|
|
|
|
Male |
n=13 (50%) |
49.1% |
ï£2=0.040 |
|
Female |
n=13 (50%) |
50.9% |
d.f.=1, p=0.8414 |
|
Race / Ethnicity |
|
|
|
|
White |
n=9 (34.6%) |
41.4% |
ï£2=1.488 |
|
Minority (Hispanic) |
n=17 (65.4%) |
58.6% |
d.f.=1, p=0.2225 |
The qPCR screening results revealed that the average amount of detectable S. wiggsiae in saliva decreased post-sealant placement (Figure 1). More specifically, these data suggest that the Pre-sealant average (Fig. 1A: 11.68 U/uL) was reduced among the Post-sealant samples (Fig. 1B. 11.14 U/uL). In addition to the average decrease in microbial sample averages, almost one-third of the samples analyzed (n=8) were found to exhibit an increase in detectable SW, while almost one-third (n=8) exhibited a decrease in detectable SW – demonstrating an overall reduction in the number of samples found harboring SW in the post-sealant samples in comparison to the pre-sealant samples.
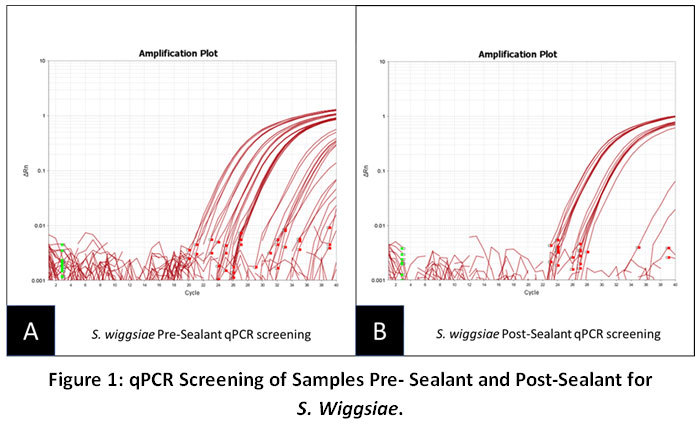 |
Figure 1: qPCR Screening of Samples Pre- Sealant and Post-Sealant for S. Wiggsiae. Click here to view Figure |
These data demonstrated an overall reduction in SW among Post-sealant samples (11.14 U/uL) compared with Pre-sealant samples (11.68 U/uL), as well as a reduction in the overall number of samples harboring S. wiggsiae (n=8).
For comparison, qPCR screening was also performed to determine the average amount of detectable S. mutans in saliva pre- and post-sealant placement (Figure 2). These data suggest that the Pre-sealant average (Fig. 2A: 14.54 U/uL) was slightly increased among the Post-sealant samples (Fig. 2B. 15.67 U/uL). In addition to the average increase in microbial sample averages, half of the samples analyzed (n=13) exhibited slight increases in SM levels, while nearly one-third (n=8) were found to exhibit slight decreases in SM levels in the post-sealant samples in comparison to the pre-sealant samples.
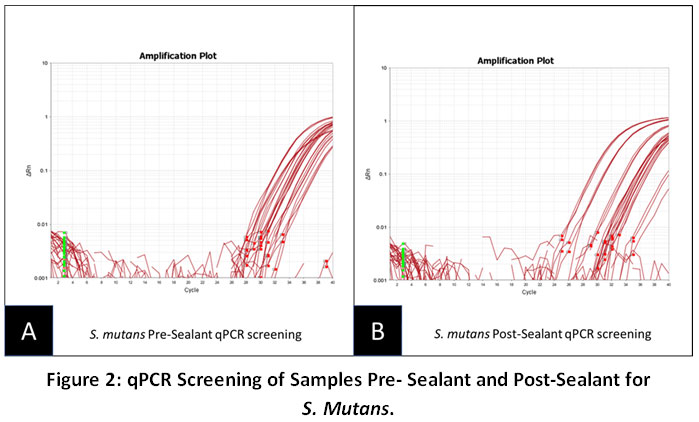 |
Figure 2: qPCR Screening of Samples Pre-Sealant and Post-Sealant for S. Mutans. Click here to view Figure |
The Pre-sealant average amount of SM (14.54 U/uL) was slightly increased among the Post-sealant samples (15.67 U/uL). In addition half of the samples analyzed (n=13) exhibited slight increases in SM levels, while nearly one-third (n=8) were found to exhibit slight decreases in SM levels.
Discussion
The primary objective of this study was to determine the effects dental sealants have on the overall quantity of S. mutans and S. wiggsiaein the oral biome in pediatric patients. The data obtained shows that there is a decrease in S. wiggsiae population post-sealant but an increase in S. mutans after a 90 day period. The decrease in S. wiggsiaecould be due to the fact that the sealants created a dysbiosis of the microbial community which competitively limited its survival. A previous study by this group has suggested that Scardovia species may inhibit the growth of some species while also enhancing the growth of others14. In fact, since the two species are not exclusively competitive, the decrease in S. wiggsiae may have facilitated the growth of S. mutans.
Additionally, not all microbiota of pathogenic bacteria have similar functions within the oral cavity15. For example,S. mutans may have initiated and facilitated the acidogenic role among the post-sealant samples, which may have contributed to the observed decreases in S. wiggsiae. These results demonstrate dental sealants on select teeth may, in fact, disrupt colonization of the oral environment by some Gram positive bacteria. However, it should also be noted that other factors, such as diet and oral hygiene, also vary from patient to patient. Any change in diet or oral care post-sealant could have increased caries risk and may partially explain the observed increases in S. mutansamong some patients.
However, more research needs to be conducted to determine the specific mechanisms responsible for these observations. Some suggestions for future research methods could be to obtain direct samples from the teeth receiving sealants instead of retrieving whole saliva samples. For example, retrieving samples from the cavosurface margins of the sealants could produce more accurate results from the sealant itself. This would provide more a method for more direct comparisons with other studies of bacterial levels and dental sealants and could limit other contributing factors such as new caries lesions from teeth that do not have sealants placed16,17. In addition, follow up data collection would also be specific to those isolated sealant placements.
Other limitations include the overall sample size and collection parameters, which might skew the findings because patient samples were all collected from one dental school and only allowed for a limited number of samples to be collected and analyzed. In addition, patient non-compliance may be another issue to be addressed, as many patients fail to return for post-sealant check up appointments – thereby limiting the number of samples that can be analyzed pre- and post-sealant placement. Finally, the small quantity of saliva among pediatric patients may also limit the overall quality and quantity of DNA obtained for this type of qPCR analysis.
Conclusions
The data from this study may suggest that dental sealants have an impact on certain cariogenic pathogens in the oral microbiota. Whether or not the impact is positive or not in light of the increase in S. mutans remains an observation that should be further analyzed. More research is required to address the limitations found in this study in order to localize the exact effect on the teeth with dental sealants.
Acknowledgements
The authors would like to thank Dr. Cody Hughes and the Department of Advanced Education, Pediatric program as well as Dr. Jeffrey Ebersole and the Office of Research at the University of Nevada, Las Vegas – School of Dental Medicine for funding and support to complete this project.
References
- Aida Bianco, LeonzioFortunato, Carmelo Giuseppe Angelo Nobile, Maria Pavia, Prevalence and determinants of oral impacts on daily performance: results from a survey among school children in Italy, European Journal of Public Health, Volume 20, Issue 5, October 2010, Pages 595–600, https://doi.org/10.1093/eurpub/ckp179.
CrossRef - Ng, M. W., Chase, I. (2013). Early Childhood Caries.Dental Clinics of North America, 2013-01-01, Volume 57, Issue 1, Pages 1-16, Copyright © 2013 Elsevier Inc.
CrossRef - Chen, K. J., Gao, S. G., Duangthip, D., Lo, E. C. M., Chu, C. H. (2018). Prevalence of early childhood caries among 5-year-old children: A systematic review. Journal of Investigative and Clinical Dentistry (10)1.https://doi.org/10.1111/jicd.12376.
CrossRef - Cui, T., Luo, W., Xu, L., Yang, B., Zhao, W., Cang, H. Progress of Antimicrobial Discovery Against the Major Cariogenic Pathogen Streptococcus mutans. Curr Issues Mol Biol. 2019 Jun 5;32:601-64 doi: 10.21775/cimb.032.601.
CrossRef - Quan, K., Kingsley, K. Effect of Dental Sealants on Oral Microbial Burden of Scardoviawiggsiae within a Pediatric Population: A Pilot Study. Microbiology Research Journal International. 24(6): 1-10, 2018; Article no.MRJI.42947.
CrossRef - Aas, J. A., Paster, B. J., Stokes, L. N., Olsen, I., & Dewhirst, F. E. (2005). Defining the normal bacterial flora of the oral cavity.Journal of clinical microbiology, 43(11), 5721–5732. doi:10.1128/JCM.43.11.5721-5732.2005.
CrossRef - Kressirer, C. A., Chen, T., Lake Harriman, K., Frias-Lopez, J., Dewhirst, F. E., Tavares, M. A., & Tanner, A. C. (2018). Functional profiles of coronal and dentin caries in children. Journal of oral microbiology, 10(1), 1495976. doi:10.1080/20002297.2018.1495976.
CrossRef - Colombo, N. H., Kreling, P. F., Ribas, L. F. F., Pereira, J. A., Kressirer, C. A., Klein, M. I., Tanner, A. C. R., Duque, C. Quantitative assessment of salivary oral bacteria according to the severity of dental caries in childhood. Arch Oral Biol. 2017 Aug 16; 83: 282–288. Published online 2017 Aug 16. doi: 10.1016/j.archoralbio.2017.08.006.
CrossRef - Kirthiga M, Murugan M, Saikia A, KirubakaranR.Risk Factors for Early Childhood Caries: A Systematic Review and Meta-Analysis of Case Control and Cohort Studies. Pediatr Dent. 2019 Mar 15;41(2):95-112.
- Junger, M. L., Griffin, S. O., Lesaja, S., & Espinoza, L. (2019). Awareness Among US Adults of Dental Sealants for Caries Prevention. Preventing chronic disease, 16, E29. doi:10.5888/pcd16.180398.
CrossRef - Wright JT, Crall JJ, Fontana M, Gillette EJ, Nový BB, Dhar V, et al. Evidence-based clinical practice guideline for the use of pit-and-fissure sealants: a report of the American Dental Association and the American Academy of Pediatric Dentistry. J Am Dent Assoc 2016;147(8):672–682.e12. 10.1016/j.adaj.2016.06.001.
CrossRef - Hurley, E., Barrett, M., Kinirons, M., Whelton, H., Ryan, C. A., Stanton, C., … O'Toole, P. W. (2019). Comparison of the salivary and dentinal microbiome of children with severe-early childhood caries to the salivary microbiome of caries-free children.BMC oral health, 19(1), 13.doi:10.1186/s12903-018-0693-1.
CrossRef - Vacharaska, A., Suvansopee, P., Opaswanich, N., Sukarawan, W. (2015) PCR detection of Scardoviawiggsiae in combination with Streptococcus mutans for early childhood cariesâ€risk prediction. European Journal of Oral Sciences.123(5).https://doi.org/10.1111/eos.12208.
CrossRef - McDaniel, S., McDaniel, J., Tam, A., Kingsley, K., Howard, K. M. (2017) Oral Microbial Ecology of Selenomonasnoxia and Scardoviawiggsiae. Microbiology Research Journal International, 21(3): 1-8. doi: 10.9734/MRJI/2017/36110.
CrossRef - Tanner, A. C. R., Kressirer, C. A., Rothmiller, S., Johansson, I., & Chalmers, N. I. (2018). The Caries Microbiome: Implications for Reversing Dysbiosis. Advances in Dental Research, 29(1), 78–85. https://doi.org/10.1177/0022034517736496.
CrossRef - Amin HE. Clinical and antibacterial effectiveness of three different sealant materials. J Dent Hyg. 2008 Fall;82(5):45. Epub 2008 Oct 1. PMID: 19055885.
- Oong EM, Griffin SO, Kohn WG, Gooch BF, Caufield PW. The effect of dental sealants on bacteria levels in caries lesions: a review of the evidence. J Am Dent Assoc. 2008 Mar;139(3):271-8; quiz 357-8. Review. PMID: 18310731.
CrossRef
.jpg)




